Adameyko Lab
Medical University of Vienna
Karolinska Institutet
Aims of research
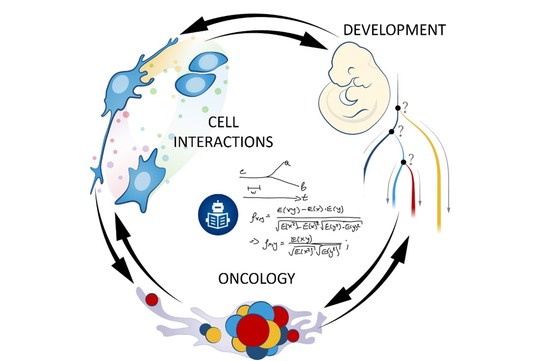
We are known for running projects that tackle different aspects of organismal biology. We look at a range of live systems from a mechanistic point of view, trying to reverse engineer different aspects of multicellular life. Tracing the incremental advancements in development of multicellular organisms from a single cell perspective allows better understanding of the complexity of the entire organism or organ system in a final phase. That is why our main strength is developmental biology. The knowledge gained from developmental biology research is widely applied in regenerative medicine. Thus, we hope to improve human health via discovering new fundamental ideas about how development, stem cells, and regeneration work.
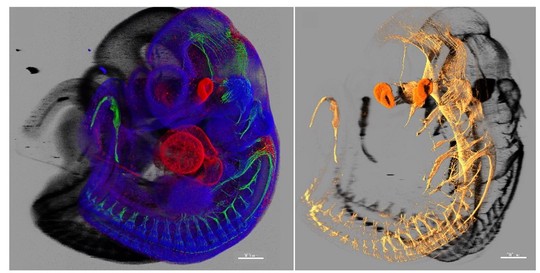
Our laboratory advances a broad spectrum of projects related to developmental biology, stem cells, EvoDevo and regenerative medicine. The methodology includes classical developmental biology approaches blended with single cell transcriptomics, 2D sequencing and 3D-reconstructions of tissues and organs based on optical or X-ray methods (micro-CT, synchrotron).
The neural crest stem cells is our primary model system, where we address general principles of cell fate choice, transcriptional and epigenetic control of a lineage progression, morphogenesis, and tissue shaping.
Lab competences
Developmental studies
Lineage tracing
Cell type ablation
Organoid and assembloid models
Advanced imaging
3D microscopy
MERFISH and Slide-seq
RNAscope
Advanced bioinformatics
scRNA-seq and scATAC-seq
Trajectory analysis and viral barcoding
Machine learning for cell fates tracing
Projects
*Featured Publications
Schwann cell precursors represent a neural crest-like state with biased multipotency
Schwann cell precursors (SCPs) are nerve-associated progenitors that can generate myelinating and non-myelinating Schwann cells but also are multipotent like the neural crest cells from which they originate. SCPs are omnipresent along outgrowing peripheral nerves throughout the body of vertebrate embryos. By using single-cell transcriptomics to generate a gene expression atlas of the entire neural crest lineage, we show that early SCPs and late migratory crest cells have similar transcriptional profiles characterised by a multipotent “hub” state containing cells biased towards traditional neural crest fates. SCPs keep diverging from the neural crest after being primed towards terminal Schwann cells and other fates, with different subtypes residing in distinct anatomical locations. Functional experiments using CRISPR-Cas9 loss-of-function further show that knockout of the common “hub” gene Sox8 causes defects in neural crest-derived cells along peripheral nerves by facilitating differentiation of SCPs towards sympathoadrenal fates. Finally, specific tumour populations found in melanoma, neurofibroma and neuroblastoma map to different stages of SCP/Schwann cell development. Overall, SCPs resemble migrating neural crest cells that maintain multipotency and become transcriptionally primed towards distinct lineages.
Single-cell transcriptomics of human embryos identifies multiple sympathoblast lineages with potential implications for neuroblastoma origin
Characterization of the progression of cellular states during human embryogenesis can provide insights into the origin of pediatric diseases. We examined the transcriptional states of neural crest– and mesoderm-derived lineages differentiating into adrenal glands, kidneys, endothelium and hematopoietic tissue between post-conception weeks 6 and 14 of human development. Our results reveal transitions connecting the intermediate mesoderm and progenitors of organ primordia, the hematopoietic system and endothelial subtypes. Unexpectedly, by using a combination of single-cell transcriptomics and lineage tracing, we found that intra-adrenal sympathoblasts at that stage are directly derived from nerve-associated Schwann cell precursors, similarly to local chromaffin cells, whereas the majority of extra-adrenal sympathoblasts arise from the migratory neural crest. In humans, this process persists during several weeks of development within the large intra-adrenal ganglia-like structures, which may also serve as reservoirs of originating cells in neuroblastoma.
Spatiotemporal structure of cell fate decisions in murine neural crest
Neural crest cells are embryonic progenitors that generate numerous cell types in vertebrates. With single-cell analysis, we show that mouse trunk neural crest cells become biased toward neuronal lineages when they delaminate from the neural tube, whereas cranial neural crest cells acquire ectomesenchyme potential dependent on activation of the transcription factor Twist1. The choices that neural crest cells make to become sensory, glial, autonomic, or mesenchymal cells can be formalized as a series of sequential binary decisions. Each branch of the decision tree involves initial coactivation of bipotential properties followed by gradual shifts toward commitment. Competing fate programs are coactivated before cells acquire fate-specific phenotypic traits. Determination of a specific fate is achieved by increased synchronization of relevant programs and concurrent repression of competing fate programs.
Recent Publications
Directionality of developing skeletal muscles is set by mechanical forces
Nature Communications,
(2023)
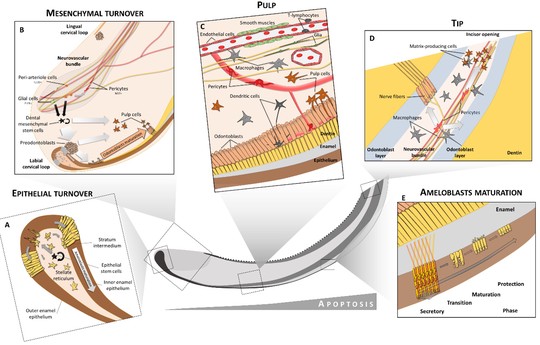
Plasticity of Dental Cell Types in Development, Regeneration, and Evolution
Journal of Dental Research,
(2023)
Adameyko Lab
Medical University of Vienna
Karolinska Institutet
Aims of research
We are known for running projects that tackle different aspects of organismal biology. We look at a range of live systems from a mechanistic point of view, trying to reverse engineer different aspects of multicellular life. Tracing the incremental advancements in development of multicellular organisms from a single cell perspective allows better understanding of the complexity of the entire organism or organ system in a final phase. That is why our main strength is developmental biology. The knowledge gained from developmental biology research is widely applied in regenerative medicine. Thus, we hope to improve human health via discovering new fundamental ideas about how development, stem cells, and regeneration work.
Our laboratory advances a broad spectrum of projects related to developmental biology, stem cells, EvoDevo and regenerative medicine. The methodology includes classical developmental biology approaches blended with single cell transcriptomics, 2D sequencing and 3D-reconstructions of tissues and organs based on optical or X-ray methods (micro-CT, synchrotron).
The neural crest stem cells is our primary model system, where we address general principles of cell fate choice, transcriptional and epigenetic control of a lineage progression, morphogenesis, and tissue shaping.
Lab competences
Developmental studies
Lineage tracing
Cell type ablation
Organoid and assembloid models
Advanced imaging
3D microscopy
MERFISH and Slide-seq
RNAscope
Advanced bioinformatics
scRNA-seq and scATAC-seq
Trajectory analysis and viral barcoding
Machine learning for cell fates tracing
Projects
Featured Publications
Schwann cell precursors represent a neural crest-like state with biased multipotency
Schwann cell precursors (SCPs) are nerve-associated progenitors that can generate myelinating and non-myelinating Schwann cells but also are multipotent like the neural crest cells from which they originate. SCPs are omnipresent along outgrowing peripheral nerves throughout the body of vertebrate embryos. By using single-cell transcriptomics to generate a gene expression atlas of the entire neural crest lineage, we show that early SCPs and late migratory crest cells have similar transcriptional profiles characterised by a multipotent “hub” state containing cells biased towards traditional neural crest fates. SCPs keep diverging from the neural crest after being primed towards terminal Schwann cells and other fates, with different subtypes residing in distinct anatomical locations. Functional experiments using CRISPR-Cas9 loss-of-function further show that knockout of the common “hub” gene Sox8 causes defects in neural crest-derived cells along peripheral nerves by facilitating differentiation of SCPs towards sympathoadrenal fates. Finally, specific tumour populations found in melanoma, neurofibroma and neuroblastoma map to different stages of SCP/Schwann cell development. Overall, SCPs resemble migrating neural crest cells that maintain multipotency and become transcriptionally primed towards distinct lineages.
Single-cell transcriptomics of human embryos identifies multiple sympathoblast lineages with potential implications for neuroblastoma origin
Characterization of the progression of cellular states during human embryogenesis can provide insights into the origin of pediatric diseases. We examined the transcriptional states of neural crest– and mesoderm-derived lineages differentiating into adrenal glands, kidneys, endothelium and hematopoietic tissue between post-conception weeks 6 and 14 of human development. Our results reveal transitions connecting the intermediate mesoderm and progenitors of organ primordia, the hematopoietic system and endothelial subtypes. Unexpectedly, by using a combination of single-cell transcriptomics and lineage tracing, we found that intra-adrenal sympathoblasts at that stage are directly derived from nerve-associated Schwann cell precursors, similarly to local chromaffin cells, whereas the majority of extra-adrenal sympathoblasts arise from the migratory neural crest. In humans, this process persists during several weeks of development within the large intra-adrenal ganglia-like structures, which may also serve as reservoirs of originating cells in neuroblastoma.
Spatiotemporal structure of cell fate decisions in murine neural crest
Neural crest cells are embryonic progenitors that generate numerous cell types in vertebrates. With single-cell analysis, we show that mouse trunk neural crest cells become biased toward neuronal lineages when they delaminate from the neural tube, whereas cranial neural crest cells acquire ectomesenchyme potential dependent on activation of the transcription factor Twist1. The choices that neural crest cells make to become sensory, glial, autonomic, or mesenchymal cells can be formalized as a series of sequential binary decisions. Each branch of the decision tree involves initial coactivation of bipotential properties followed by gradual shifts toward commitment. Competing fate programs are coactivated before cells acquire fate-specific phenotypic traits. Determination of a specific fate is achieved by increased synchronization of relevant programs and concurrent repression of competing fate programs.